Introduction to LC-MS Part3
Part3
Development of the atmospheric pressure ionization (API) method made it possible to ionize a wide range of organic compounds, which has expanded the range of applications for LC-MS analysis. By the way, what kind of mass spectra are obtained using API? That is the topic of this page.
Mass Spectra from API
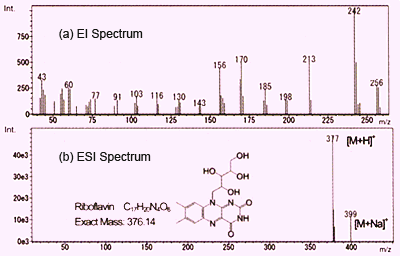
Fig. 1 Mass Spectra of Vitamin B2
As described before, both ESI and APCI result in the detection of primarily protonated molecules and also metal or solvent adduct ions. On this page, we compare these methods with electron ionization (EI) which is commonly used in GC-MS. Figure 1 shows spectra for vitamin B2 (riboflavin) obtained using EI and ESI. These are graphed with ion intensity on the vertical axis and mass-to-charge ratio (m/z) on the horizontal axis.
EI uses an electron beam to liberate one electron from molecules in a gas phase to create molecular ions (radical cations). These instantly burst to generate a group of fragment ions. Structural information can be obtained by considering the pattern of these fragment ions. However, in many cases, no molecular ion peeks are detected. In the case of Figure 1a, no molecular ions are detected, but rather, only fragment ions are detected. Obtaining molecular mass information is difficult using EI, which requires using a complementary analytical method such as chemical ionization (CI).
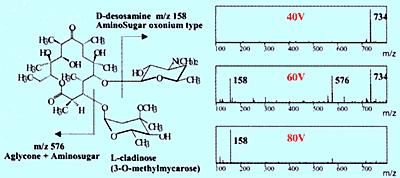
Fig. 2 Collision-Induced Dissociation Spectra of Erythromycin
In contrast, using ESI, which is a soft ionization method, provides a simple spectrum with a protonated molecule detected at m/z 377 and a sodium adduct ion detected at m/z 399, and almost no fragment ions. In this way, molecular mass information, which is important for predicting the structure of unknown compounds, can be obtained easily using API. (In this example, since vitamin B2 has a basic functional group, the positive ion mode is used.) However, because use of API tends not to produce fragment ions, it seems difficult to obtain structural information of functional groups and others by analyzing the fragment ions.
Nevertheless, structural information can be obtained using API if using a method called collision-induced dissociation (CID) to create fragment ions, then measuring those fragment ions. CID can occur at the electrostatic lens area (Figure 2) or using a tandem type mass spectrometer equipped with a collision chamber, which is described below. Figure 2 shows an example of generating fragment ions from an erythromycin antibiotic by increasing the voltage applied to the electrostatic lens. These ions can be used to identify the minor components that appear in the chromatogram (refer to Shimadzu Application News, LCMS No. C21).
Measuring Multivalent Ions
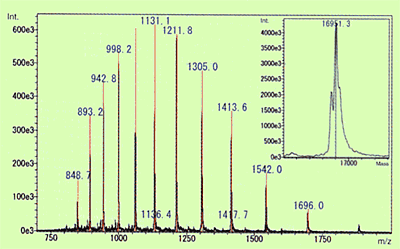
Fig. 3 Molecular Mass of Myoglobin in Equine Heart Muscle Calculated Using ESI Spectrum and Deconvolution
Of the API methods, use of ESI in particular, is known to sometimes generate molecular ions with multiple charges for compounds that have multiple potential ionization points. (In the case of cations, this means multiple protons are added [M+nH]n+.) The level of protonation is strongly influenced by the pKa level of the compound or pH level of the solution. When this type of multivalent ion is observed, molecular mass information can be obtained even for compounds with a molecular mass that exceeds the measurement range of the mass spectrometer. Therefore, this is used to measure extremely large and highly-polar biological macromolecules, such as proteins and nucleic acids. Figure 3 shows an ESI spectrum of equine myoglobin as an example of protein. In this case, ions with valences from 9 to 20 were detected and molecular mass was calculated by deconvolution to be 16951.3. Compared to the theoretical molecular mass, 16951.5, calculated from the composition of amino acids, the error was less than 0.002 %, which indicates that an extremely accurate value was obtained.
This is a big difference from GC-MS, which usually only provides peaks for z=1.


