Oct 20, 2020
In a joint research effort, the team of Mari Tohya, Ph.D., at Department of Microbiology headed by Professor Teruo Kirikae of Juntendo University School of Medicine, and Koichi Tanaka Mass Spectrometry Research Laboratory has proposed three strains of Pseudomonas oleovorans isolated from swabs of inpatient wounds in Myanmar as a novel species. According to whole genome data and fatty acid contents, these three strains were found to be closely related type strains of P. guguanensis and P. mendocina within the P. oleovorans group, but to be a novel species, for which we name Pseudomonas yangonensis sp. nov.
The results of this study were published in the International Journal of Systematic and Evolutionary Microbiology 2020, 70, 3597 -3605.
Study Overview
The genus Pseudomonas is a large taxon with multiple strains which are widely distributed in natural environments. Most of them are not pathogens in plants or animals and are not directly associated with disease. However, a few species are known to cause infections in humans. For example, P. oleovorans subsp. oleovorans and P.mendocina have been reported to be isolated from patients, unlike other species in this group.
Through a survey of carbapenem-resistant Gram-negative pathogens in medical settings in Myanmar, these three strains were isolated from wound swabs of three patients. Analysis of their biochemical properties used to result in their tentative identification as P. putida. Following further analysis and characterization including whole-genome comparisons confirmed that these strains were a distinct species in the P. oleovorans group.
A comparison of the MALDI mass spectra of the three strains and several related strains revealed four peaks derived from the proteins that characterized the three strains.
The peak lists obtained from the mass spectra were evaluated by principal component analysis (PCA) using Shimadzu’s statistical analysis software eMSTAT Solution™. According to the score plot of PCA, the three strains formed a cluster which is separated from clusters of other type strains.
Protein analysis by mass spectrometry indicated that these three strains are a novel species.
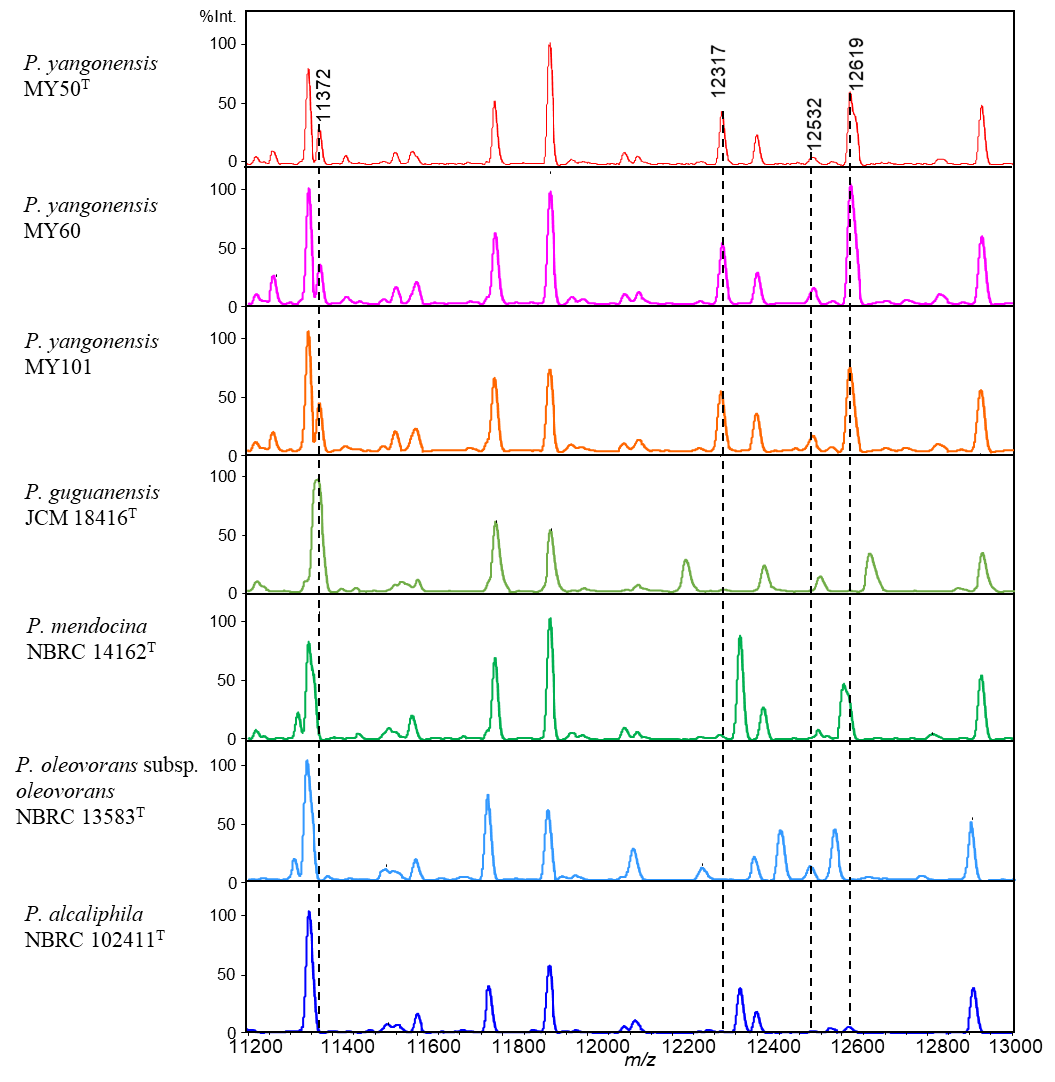
Fig. 1. Comparative MALDI-TOF MS profiles (m/z 11200–13000) of a novel species (a) P. yangonensis MY50T, (b) P. yangonensis MY60, (c) P. yangonensis MY101 and (d, e, f, g) related strains of the P. oleovorans group. The peaks at m/z 11372, 12317,12532 and 12619 were features specific to MY50T, MY63 and MY101, distinguishing them from the other related strains. Reprinted from: International Journal of Systematic and Evolutionary Microbiology, 70, 6, 3597-3605, 2020
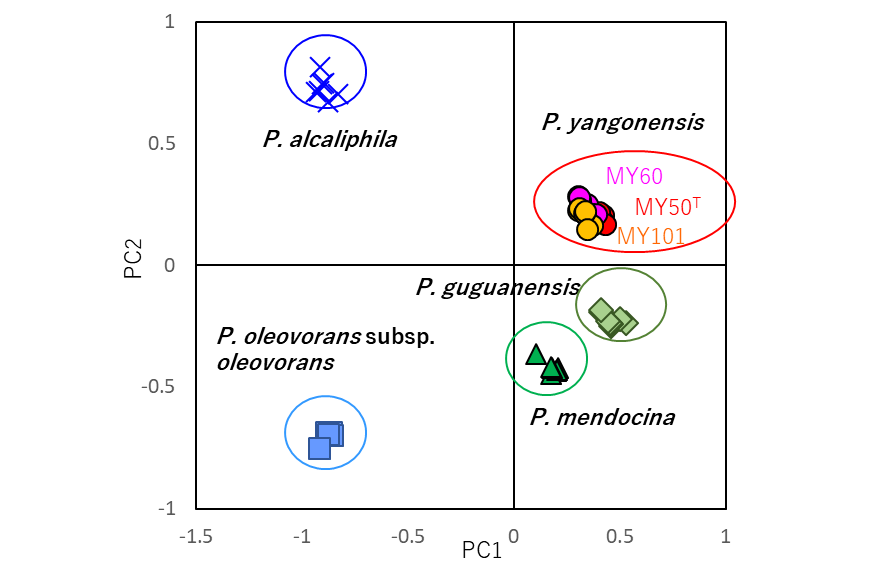
Fig. 2. PCA score plots constructed by eMSTAT Solution indicated that yangonensis MY50T, P. yangonensis MY60, P. yangonensis MY101 formed a cluster which is separated from clusters of other type strains. Reprinted from: International Journal of Systematic and Evolutionary Microbiology, 70, 6, 3597-3605, 2020
【Paper Information】:
Mari Tohya, Shin Watanabe, Kanae Teramoto, Tatsuya Tada, Kyoko Kuwahara-Arai, San Mya, Khwar Nyo Zin, Teruo Kirikae, Htay Htay Tin
“Pseudomonas yangonensis sp. nov., isolated from wound samples of patients in a hospital in Myanmar”
International Journal of Systematic and Evolutionary Microbiology, 70, 6, 3597-3605, 2020
DOI: 10.1099/ijsem.0.004181


