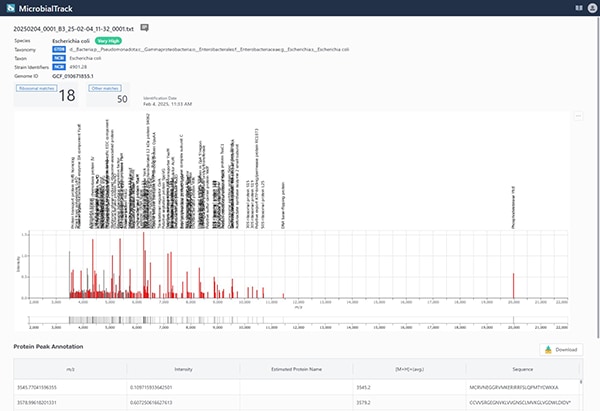
May 20, 2025 | News & Notices Shimadzu Corporation launches "MicrobialTrack" – a new global platform for microbial identification powered by the world’s largest database.
Announcing a next-generation platform for MALDI-TOF MS-based microbial identification, featuring the most extensive database that covers approximately 85,000 prokaryotic species, including not only well-known, previously described microorganisms but also hard-to-culture and uncultured microorganisms — MicrobialTrack redefines the boundaries of MALDI-TOF MS-based microbial identification.
Shimadzu Corporation has launched MicrobialTrack, an innovative software platform designed for high-precision microbial identification across a wide range of fields, including food safety, healthcare, drug discovery, and environmental research. The software leverages a proprietary, large-scale protein mass database built from publicly available microbial genomic information. It analyzes measurement data obtained through Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS). The MicrobialTrack database features a comprehensive collection of known microbial entries, comprising approximately 85,000 prokaryotic species (Bacteria and Archaea), including those with validly published taxonomic names as well as difficult-to-culture and previously uncultured microorganisms. This significantly expands the scope of identification beyond the limits of conventional methods.
In recent years, the demand for identifying microorganisms has increased worldwide due to heightened hygiene awareness, the proliferation of chilled foods, and the emergence of antibiotic-resistant bacteria. In clinical and food microbiology testing, the "fingerprint method" identifies microorganisms based on measurement results obtained from MALDI-TOF MS. This method provides results in approximately three hours, including the pre-treatment of microbial isolates (cells), MALDI-TOF MS measurement, and data analysis; however, the database is generally restricted to approximately 5,000 well-known species, which poses challenges for identification accuracy and limits its applications in a wide range of fields in microbiology.
To overcome this limitation, MicrobialTrack employs a "proteomics approach" to identify microbial species based on the similarity between the theoretical masses of proteins estimated from genomes and the observed masses of proteins derived from samples. This product utilizes the theoretical mass database for prokaryotic microorganisms, "GPMsDB," which is based on approximately 400,000 genome entries obtained from public genome databases. The analysis targets approximately 85,000 prokaryotic species predicted based on genomic information, including difficult-to-culture and uncultured microorganisms. This capability allows for the identification of a wide range of microorganisms that could not be identified using the fingerprint method with MALDI-TOF MS.
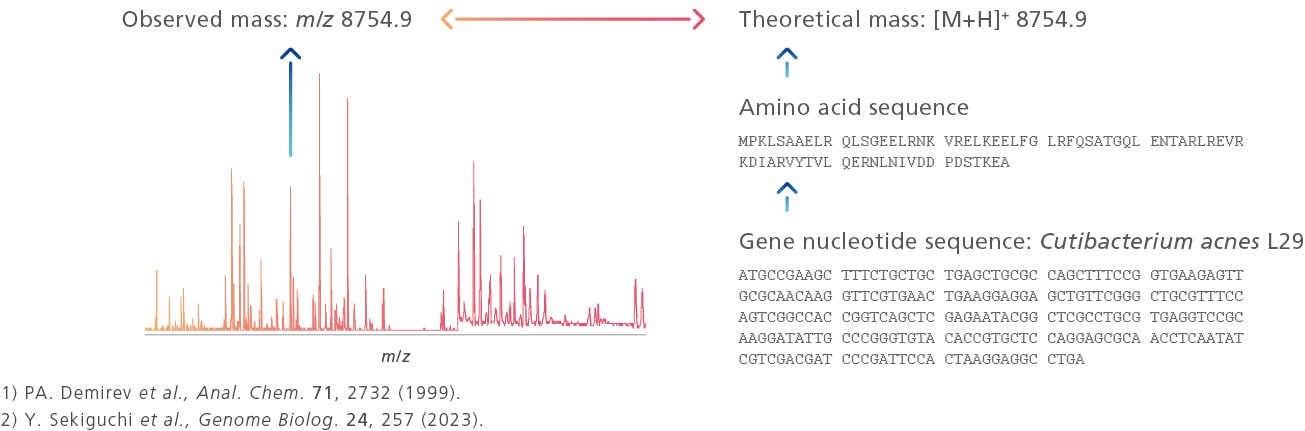
Figure:Principles of microorganism identification by proteomics-based approach
MicrobialTrack is a cloud-based service that enables the identification of microorganisms in a web browser. The service does not require a dedicated PC or software installation. Users simply register and then access the designated URL to proceed with the analysis. Several hundred thousand new prokaryotic genome sequences are published yearly and added to public databases. The MicrobialTrack database is updated regularly to include newly added taxa as genome sequences and to reflect the most recent taxonomic system, which is continually evolving. Users will have access to the latest MicrobialTrack database during the license period.
MicrobialTrack is a product developed through joint research with the National Institute of Advanced Industrial Science and Technology (AIST) and the National Institute of Technology and Evaluation. In 2023, AIST and our company published the basis of this microorganism identification technology, including the genomically predicted mass database "GPMsDB" and an original algorithm for analyzing mass spectra.1) MALDI, the technology that earned our Executive Research Fellow Koichi Tanaka the Nobel Prize in Chemistry, involves irradiating a sample mixed with a matrix (ionization assist agent) with a laser to ionize high-molecular compounds such as proteins without breaking them.
- 1) Y. Sekiguchi et al., Genome Biolog, 24 (2023).
For more details, visit
MicrobialTrack


