LC/MS/MS Method Package for D/L Amino Acids
For LabSolutions™ LCMS Software

Most important amino acids exist as stereoisomers. D- and L- forms of mirror image isomers, or enantiomers, are named according to their activity on polarized light. By using CROWNPAK CR-I(+) and CR-I(-) columns with chiral stationary phases, the D- and L-forms of amino acids can be analyzed separately. With CR-I(+) elution order is from D- to L-, and with CR-I(-) the elution order is reversed.
In Just Ten Minutes, Chiral Amino Acids Can Be Analyzed Simultaneously
With conventional chiral amino acid analysis, it is necessary to perform derivatization or use very long run times. With this method package, derivatization is not necessary, and high-sensitivity analysis can be performed in a short period of time, bringing efficiency to the chiral separations laboratory.
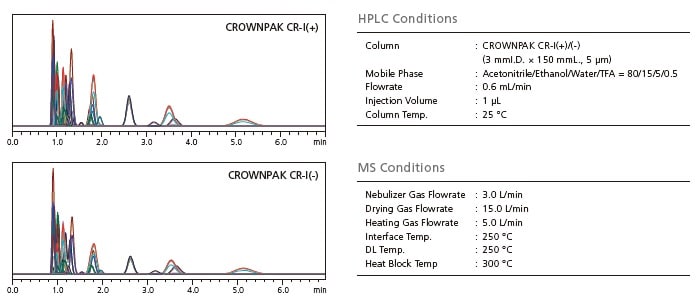
All D/L Amino Acids Can Be Quantified by Column Switching
The physicochemical properties of Glutamine and Lysine, Isoleucine and allo-Isoleucine, Threonine and allo-Threonine are extremely similar. They have virtually the same MS/MS fragmentation patterns, and share many of their MRM transitions. Chromatographic separation is therefore required to analyze them individually by LC-MS/MS. Even if there is coelution on the CR-I(+) column, confirmation can be made by automated switching to a secondary CR-I(-) column.
| Analysis of D/L-Isoleucine, D/L-allo-Isoleucine, and D/L-Leucine |
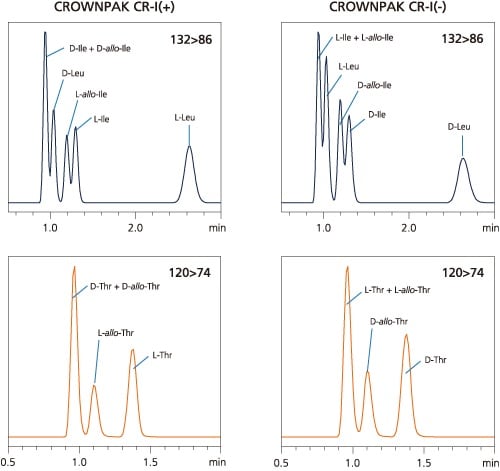 |
| Analysis of D/L-Threonine, and D/L-allo-Threonine |
List of Registered Amino Acids
- D/L-Alanine
- D/L-Arginine
- D/L-Asparagine
- D/L-Aspartic acid
- D/L-Cysteine
- D/L-Glutamine
- D/L-Glutamic acid Glycine
- Glycine
- D/L-Histidine
- D/L-Isoleucine
- D/L-allo-Isoleucine
- D/L-Leucine
- D/L-Lysine
- D/L-Methionine
- D/L-Phenylalanine
- DL-Proline
- D/L-Serine
- D/L-Threonine
- D/L-allo-Threonine
- D/L-Tryptophane
- D/L-Tyrosine
- D/L-Valine
Precautions
1. DL-Proline is a secondary amine, so it cannot be separated with these analysis conditions.
2. LabSolutions LCMS Ver. 5.86 or later is required.
The analysis method of this method package was developed based on the reserach of the Fukusaki Lab, Division of Science and Biotechnology, Graduate School of Engineering, Osaka University.
Reference: Nakano,Y., Konya,Y., Taniguchi,M., Fukusaki,E., Journal of Bioscience and Bioengineering , 123, 134-138 (2016)
LabSolutions and LCMS are trademarks of Shimadzu Corporation.
CROWNPAK is a registered trademark of Daicel Corporation.
News / Events
-
Shimadzu has released the LCMS-8065XE
The new LCMS-8065XE is a triple-quadrupole mass spectrometer with EVOLVED, EFFICIENT, and EXACT capabilities. These exceptional capabilities ensure high reliability and enhanced productivity, empowering the laboratory for the future.
-
Latest issue of Shimadzu Journal, featuring Environmental Analysis, has come out.
This issue showcases advanced technologies and research tackling the global challenges posed by PFAS. As part of Shimadzu’s ongoing commitment to sustainability and problem solving, we strive to reduce environmental impacts and build a better future.
-
New High Resolution Accurate Mass Library for Forensic Toxicology
Perform forensic toxicology screening for drugs of abuse, psychotropic drugs, pharmaceuticals, pesticides, and natural toxins using this high-resolution accurate mass database.
-
Shimadzu has released the Shim-vial™ H glass, S glass.
Shimadzu provides high-quality vials that thoroughly eliminate these risks by visually inspecting each vial, allowing them to be used with confidence.
-
INTERNATIONAL MASS SPECTROMETRY CONFERENCE 2024
Visit the Shimadzu booth at the International Mass Spectrometry Conference (IMSC) 2024.
-
Metabolomics 2024
Shimadzu Lunch Presentation at Metabolomics 2024
Date: June 20th, 2024 (Thursday)
Time: 12:25 – 1:25 p.m.




