Quality Check for PCR direct-sequencing
PCR direct sequencing is a technique in which, rather than cloning the amplification products obtained as a result of the PCR reaction, they are used directly as a template for determining the base sequence. This is a very effective method because it allows base sequence information to be obtained quickly and reduces the influence of base take-up mistakes following cloning.
However, those results may vary widely depending on the amplification situation following PCR. In order to increase that success rate, it is necessary to verify the purity (quality and quantity) of the PCR products prior to the sequencing reaction. That verification has traditionally been conducted using agarose gel electrophoresis.
However, there are cases in which the sequence data obtained in the sequence analysis indicates the possibility of closely related substrates for reaction, even when it appears that there is only a single target PCR product. One possible cause for this is the limited sensitivity of agarose gel electrophoresis. With this method, it is difficult to elucidate the existence of trace amounts of PCR side reaction products among the principal reaction products.
Here, we introduce a PCR product purity assay in a PCR direct sequencing process conducted in the search for the housekeeping genes of cruciferae plant family by the Research Institute for Biological Sciences, Okayama Prefectural Technology Center for Agriculture, Forestry, and Fisheries.
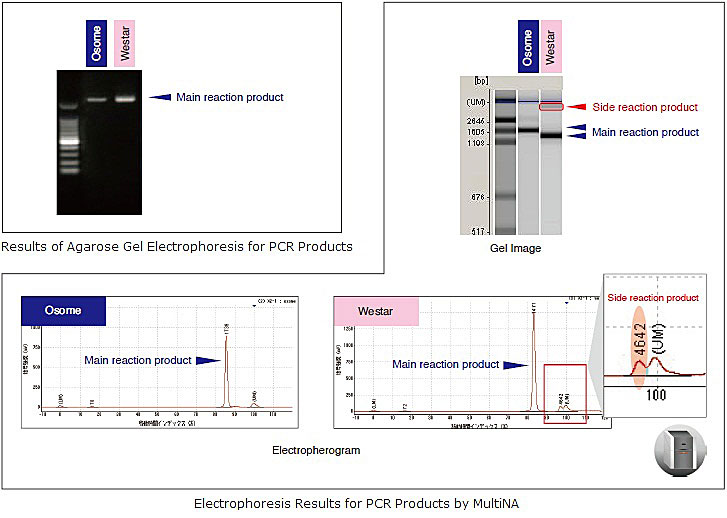
The various PCR products were detected at about the same size using agarose gel electrophoresis even though the sizes were slightly different. In the electrophoresis results obtained using the MultiNA, a side reaction product was detected in the Westar sample. This is noticeable in the gel image results, but in the electropherogram, DNA was clearly detected as peaks. However, side reaction products were not detected by agarose gel electrophoresis.
With the MultiNA, the concentration can be calculated from the area of the peak in the electropherogram. This value is sufficiently adequate for use in the sequence reaction. Thus, the MultiNA is extremely useful for conducting purity assays of PCR products as an essential component of the PCR direct sequencing process.
|
* |
Samples and data used for this study were provided by the Plant Immunology Research Group, Research Institute for Biological Sciences (RIBS), Okayama Prefectural Technology Center for Agriculture, Forestry, and Fisheries. |
Microchip Electrophoresis System for DNA/RNA

DNA and RNA samples are separated by size by the electrophoresis system using a microchip so that the size of nucleic acid (DNA/RNA) samples is verified and approximately quantitated. The microchip achieves an electrophoresis system capable of speedily conducting electrophoresis separation, and a fluorescent detector ensures that analysis is performed to high sensitivity and, moreover, fully automatically.


