MicrobialTrack - Features
MALDI-TOFMS Microbial Identification Software
Database Built from Genomic Data
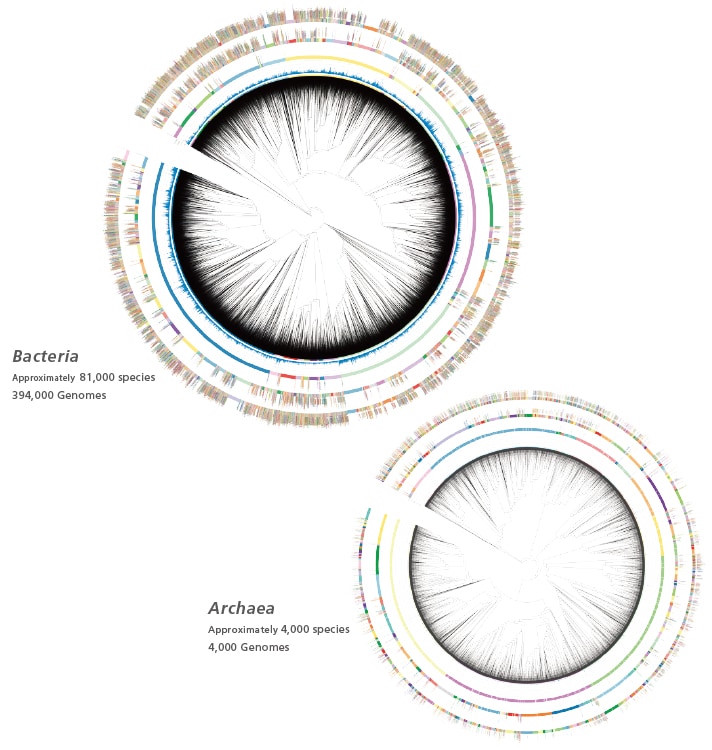
The MicrobialTrack database includes approximately 18,000 prokaryotic species (Bacteria and Archaea) with valid published taxonomic names, as well as about 85,000 prokaryotic species predicted based on genomic information, including difficult-to-culture and uncultured microorganisms. This database facilitates the identification of a wide variety of phylogenetic taxa beyond the capabilities of previous MALDI-TOF MS microorganism identification systems. Conventionally, identifying 96 cultures of isolates using nucleic acid-based techniques takes around 2 days, but MALDI-TOF MS with MicrobialTrack mass spectral analysis software can identify those 96 isolates within 3 hours Note.
Note: Throughput time for processing bacterial liquid culture using formic acid and acetonitrile.
Taxonomy Framework Based on Genomics
Identification results are displayed with taxonomic names according to the Genome Taxonomy Database (GTDB, https://gtdb.ecogenomic.org). The GTDB defines species-level taxa based on the closeness of genomic sequences using average nucleotide identity (ANI: percentage identity of homologous regions between two genome sequences) and alignment factor (AF: percentage of homologous regions shared by two genome sequences). In the GTDB, the criteria for the species level are defined as having an ANI of 95% or higher and an AF of 65% or higher. MicrobialTrack uses the same criteria for defining species-level taxa and employs the same taxonomy string as the GTDB for other taxonomic ranks.
The MicrobialTrack database contains predicted theoretical protein masses derived from genomic data of uncultured and difficult-to-culture microorganisms, such as metagenome-assembled genomes (MAGs) and single-cell assembled genomes (SAGs). Many of these uncultured microorganisms lack valid taxonomic names and are instead assigned placeholder genus or species names in the GTDB (e.g., MGIIa-L1 and sp002965065). It is important to note that these placeholder names do not qualify as valid taxonomic names. Additional information regarding taxa with placeholder names is available on the GTDB website (https://gtdb.ecogenomic.org). The designations of these taxa may change as new research uncovers further information and valid published taxonomic names are assigned.
Optimized for Different Purposes

- Users can select either of the two databases, "All" and "Reps", for microbial identification. The “All” database contains theoretical masses predicted from all genome sequences. In contrast, the “Reps” database includes those derived only from a single genome sequence representing a species-level taxon.
- For most species-level taxa, "All" contains protein information from multiple strains (genomes). For example, the taxon Escherichia coli contains protein information from over 30,000 genome sequences in the "All" database. “All” searches for the most closely matched strains in the database.
- In contrast, the “Reps” database contains only protein information from a single strain (genomic information) for one species. The “Reps” database is utilized to quickly estimate taxonomic identification at the genus and species levels. Searching within the “Reps” database allows evaluating which species closely match the measured spectrum against a list of potential candidates.
Note: The number of data points in the figure is based on information as of March 2025.
Regular Updates, Aligned with the Latest Taxonomic Framework

The MicrobialTrack database is regularly updated to introduce newly identified bacterial and archaeal taxa and to reflect the latest GTDB taxonomy framework. The database is managed on a server, ensuring users have seamless access to the latest version during the license period.
Proteomics-Based Identification
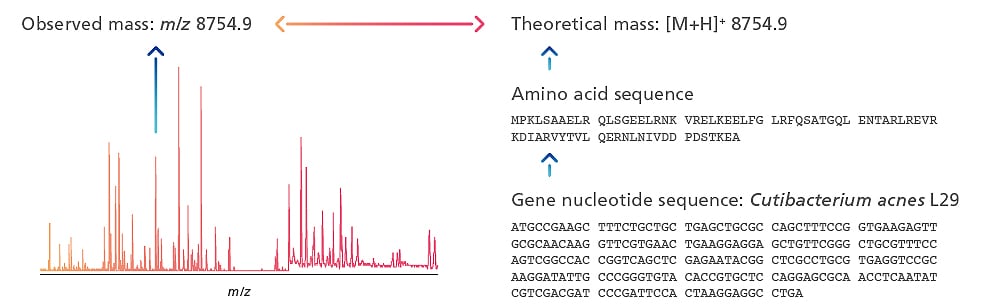
MicrobialTrack utilizes a database of theoretical masses predicted from genomic information to identify unknown microorganisms. Identification is achieved by comparing the observed masses from microbial cell MALDI-TOF MS analysis with theoretical masses. Advancements in genome sequencing technologies have greatly expanded prokaryotic genome sequence entries in public nucleotide sequence databases 1). A large-scale database of theoretical masses developed for MicrobialTrack and an original identification algorithm enable the rapid identification of a wide range of microbial species, independent of culture conditions or MALDI-TOF MS models 2).
Accessible Everywhere Via a Cloud Service
No Matter the Location, Easy to Implement
As a cloud-based service, there is no need to purchase a dedicated PC or install software. Users can access the dedicated URL from their own PC in any location as long as they have an internet-connected environment.
Robust Security
MicrobialTrack operates on a reliable cloud infrastructure with strict security features, including access controls. It maintains safety through regular monitoring of access logs, vulnerability assessments, and software updates, ensuring that the data of other users is not referenced, allowing you to use it with confidence. Furthermore, by utilizing multi-factor authentication (MFA), you can obtain an even higher level of security.

Simple Operation
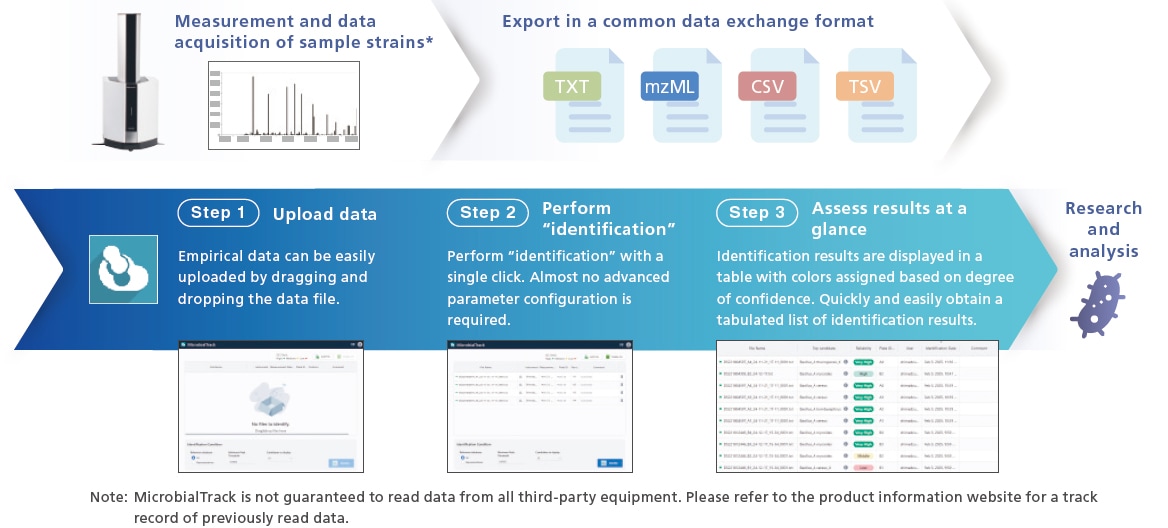
Identification Completed in Just Three Steps
Microbial cells are analyzed using MALDI-TOF MS. Since it supports a universal format, the platform is vendor neutral. The resulting peak lists are compared to a theoretical mass database to identify the microorganisms. The identification process can be completed in just three steps. The identification results present a ranked list of candidate species, along with icons that are color-coded based on the degree of confidence. The results also allow users to verify which proteins from the microbial species were detected as the peaks.
Advanced Identification of "Species Complexes"
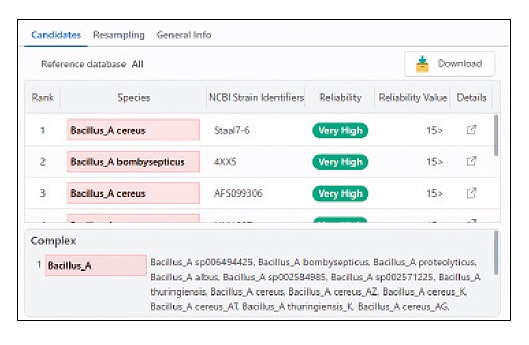
MicrobialTrack redefines the "species complex" as taxa that comprise multiple species-level taxa, which are genetically distinct but often difficult to distinguish based solely on theoretical protein masses. MicrobialTrack identifies such species complexes based on theoretical protein masses and notifies users of this information.
Multifaceted Evaluation of Identification Results
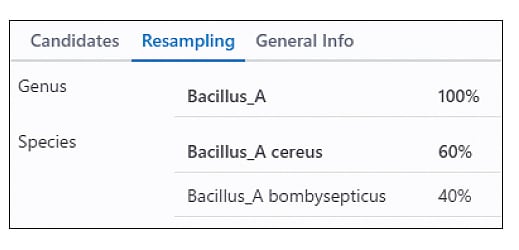
Identification results vary based on several factors, including the condition of the sample microorganisms, the preparation of the samples, and the quality of the MALDI mass spectra. Therefore, MicrobialTrack employs multiple performance metrics to evaluate each identification result from various perspectives. One of these methods involves using a portion of randomly selected observed peaks to repeat the identification process, and then displays which genus and species are identified as top-hit organisms during resampling, along with their frequency of occurrence. Based on this random resampling, the software provides an estimate of the identification certainty level at both the genus and species levels.
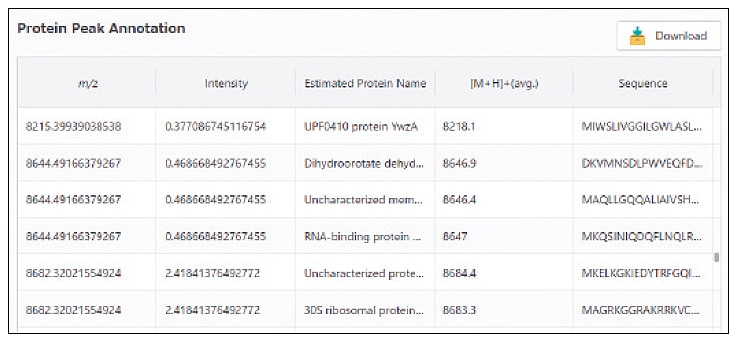
Advanced Proteomics Platform
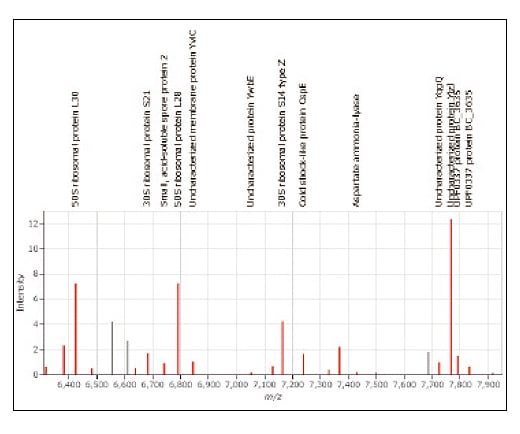
MicrobialTrack uses the theoretical masses in the database to estimate which peak corresponds to which protein. Such a list of inferred protein names is displayed on the mass spectrum and in a tabular list. Mass peaks that match theoretical protein masses are also displayed in red for easy recognition in a mass peak graph in the software. As an optional feature, MicrobialTrack also allows users to view protein names, theoretical masses, and amino acid sequences for mass peaks.