LC/MS/MS Method Package for Cell Culture Profiling Ver. 3
For LabSolutions™ LCMS Software
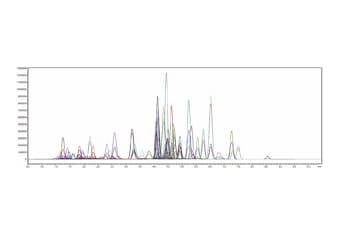
Simultaneous Analysis Conditions for 144 Compounds

Version 3 of this method package adds metabolites characteristic of CHO cells, which are frequently used in the manufacture of monoclonal antibodies. Together with existing analytes, this Method Package enables the analysis of 144 compounds in under 20 minutes per sample. Both culture supernatant components and cellular metabolites can be analyzed using this product owing to enhancement of the target analytes and optimization of measurement parameters.
This method package makes it easier to obtain more detailed cell culture profiling data.
Features
-
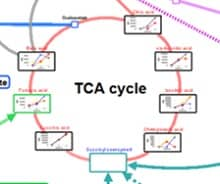
Together with existing analytes, including amino acids, vitamins, basal medium compounds,...
-
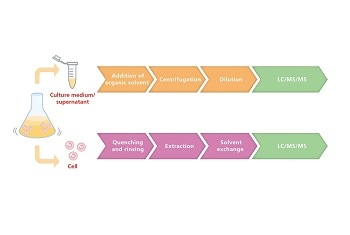
Metabolites present in both the cell medium supernatant and within cells can be analyzed,....
-
Volcano plots, principal component analysis, hierarchical clustering, and other analyses can easily be performed using....
Videos
-
Advance Cell Culture Media Analysis and Monitoring By LC-MS/MS
News / Events
-
Shimadzu has released the LCMS-8065XE
The new LCMS-8065XE is a triple-quadrupole mass spectrometer with EVOLVED, EFFICIENT, and EXACT capabilities. These exceptional capabilities ensure high reliability and enhanced productivity, empowering the laboratory for the future.
-
Latest issue of Shimadzu Journal, featuring Environmental Analysis, has come out.
This issue showcases advanced technologies and research tackling the global challenges posed by PFAS. As part of Shimadzu’s ongoing commitment to sustainability and problem solving, we strive to reduce environmental impacts and build a better future.
-
New High Resolution Accurate Mass Library for Forensic Toxicology
Perform forensic toxicology screening for drugs of abuse, psychotropic drugs, pharmaceuticals, pesticides, and natural toxins using this high-resolution accurate mass database.
-
Shimadzu has released the Shim-vial™ H glass, S glass.
Shimadzu provides high-quality vials that thoroughly eliminate these risks by visually inspecting each vial, allowing them to be used with confidence.
-
INTERNATIONAL MASS SPECTROMETRY CONFERENCE 2024
Visit the Shimadzu booth at the International Mass Spectrometry Conference (IMSC) 2024.
-
Metabolomics 2024
Shimadzu Lunch Presentation at Metabolomics 2024
Date: June 20th, 2024 (Thursday)
Time: 12:25 – 1:25 p.m.