Metabolites Method Package Suite
For LabSolutions™ LCMS and GCMSsolution™

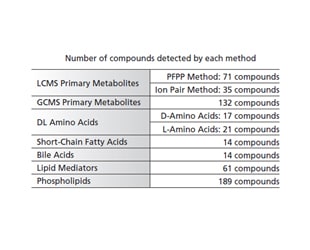
Provides ready-to-use methods for over 1900 metabolites
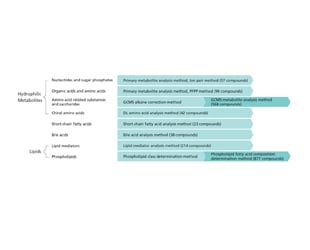
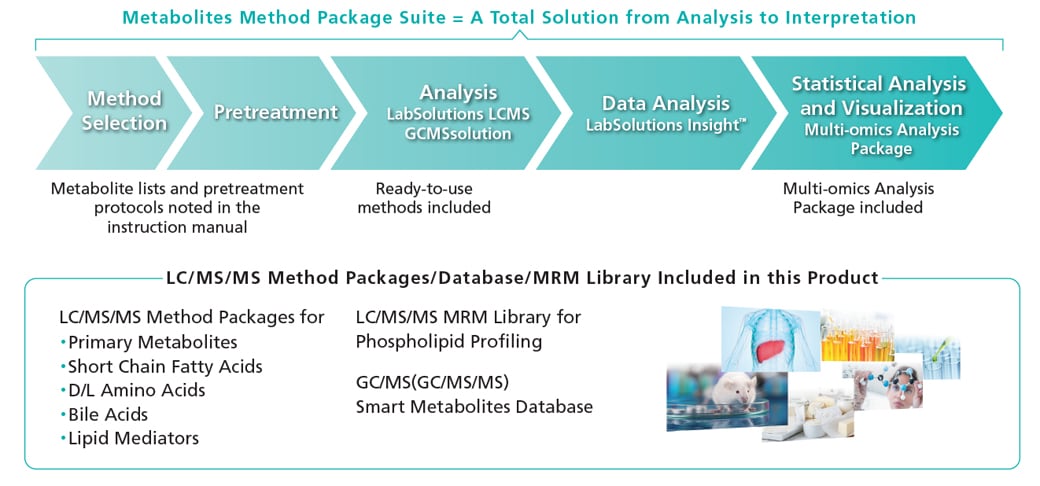
This suite allows comprehensive analysis of over 1900 metabolites without the need for investigation of separation conditions, MRM optimization or parameter settings. The range of metabolites spans both hydrophilic and hydrophobic compounds, including amino acids, short-chain fatty acids, sugars, nucleotides, bile acids, and lipids. The suite consists of five LC/MS/MS Method Packages including ready-to-use methods for the LCMS-8050/8060 series, the LC/MS/MS MRM Library for Phospholipid Profiling, the Smart Metabolites Database™ for GC/MS(GC/MS/MS), and a Multi-omics Analysis Package.
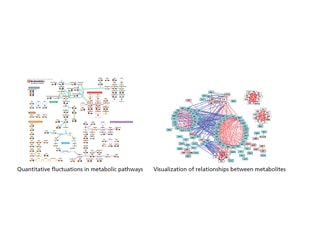
The Multi-omics Analysis Package included in this product supports not only regular analysis but also large volume data analysis and interpretation. The Multi-omics Analysis Package includes metabolic pathways and other contour maps corresponding to the Method Packages. This makes it easy to visualize fluctuations in the quantitative values of metabolites across metabolic pathways. Data filtering functions and statistical analysis can be applied to the network of compound relationships, providing a total solution for metabolite analysis.
Features
-
To start analysis without an investigation of measurement conditions, select a method suited to the analysis aims and the relevant compounds. Protocols are also included for extraction from biological tissue, plasma, and feces, as well as derivatization and other pretreatments.
-
Recently the analysis of metabolites in the field of intestinal flora has expanded in scope.
-
With the included Multi-omics Analysis Package, it is possible to display quantitative fluctuations in metabolites on a metabolic map, and to visualize relationships between metabolites.
News / Events
-
Shimadzu has released the LCMS-8065XE
The new LCMS-8065XE is a triple-quadrupole mass spectrometer with EVOLVED, EFFICIENT, and EXACT capabilities. These exceptional capabilities ensure high reliability and enhanced productivity, empowering the laboratory for the future.
-
Latest issue of Shimadzu Journal, featuring Environmental Analysis, has come out.
This issue showcases advanced technologies and research tackling the global challenges posed by PFAS. As part of Shimadzu’s ongoing commitment to sustainability and problem solving, we strive to reduce environmental impacts and build a better future.
-
New High Resolution Accurate Mass Library for Forensic Toxicology
Perform forensic toxicology screening for drugs of abuse, psychotropic drugs, pharmaceuticals, pesticides, and natural toxins using this high-resolution accurate mass database.
-
Shimadzu has released the Shim-vial™ H glass, S glass.
Shimadzu provides high-quality vials that thoroughly eliminate these risks by visually inspecting each vial, allowing them to be used with confidence.
-
INTERNATIONAL MASS SPECTROMETRY CONFERENCE 2024
Visit the Shimadzu booth at the International Mass Spectrometry Conference (IMSC) 2024.
-
Metabolomics 2024
Shimadzu Lunch Presentation at Metabolomics 2024
Date: June 20th, 2024 (Thursday)
Time: 12:25 – 1:25 p.m.